Crop wild relatives are an important reservoir of natural biodiversity. However, incorporating wild genetic diversity into breeding programs is often hampered by reproductive barriers and the lack of accurate genomic information. We assembled a high-quality, centromere accurately-anchored genome of Gossypium anomalum, a stress-tolerant wild cotton species. We provided a strategy to transfer and discover agronomically valuable genes from wild diploid species to tetraploid cotton cultivars. With a bridge parent of (G. hirsutum ×G. anomalum)2 hexaploid, we firstly developed a set of 74 diploid wild cotton species G. anomalum chromosome segment substitution lines (CSSLs) on the G. hirsutum background. This set of CSSLs included 70 homozygous substitutions and four heterozygous substitutions, and collectively contained about 72.22% of the G. anomalum genome. Twenty-four QTLs associated with plant height, yield, and fiber qualities were detected on 15 substitution segments. Integrating the reference genome with agronomic traits evaluation of CSSLs, two G. anomalum genes encoding peroxiredoxin and putative callose synthase 8 respectively conferring drought tolerance and improving fiber strength are readily located and cloned. We have demonstrated the power of a high-quality wild species reference genome in identifying agronomic valuable alleles to facilitate interspecific introgression breeding in crop.
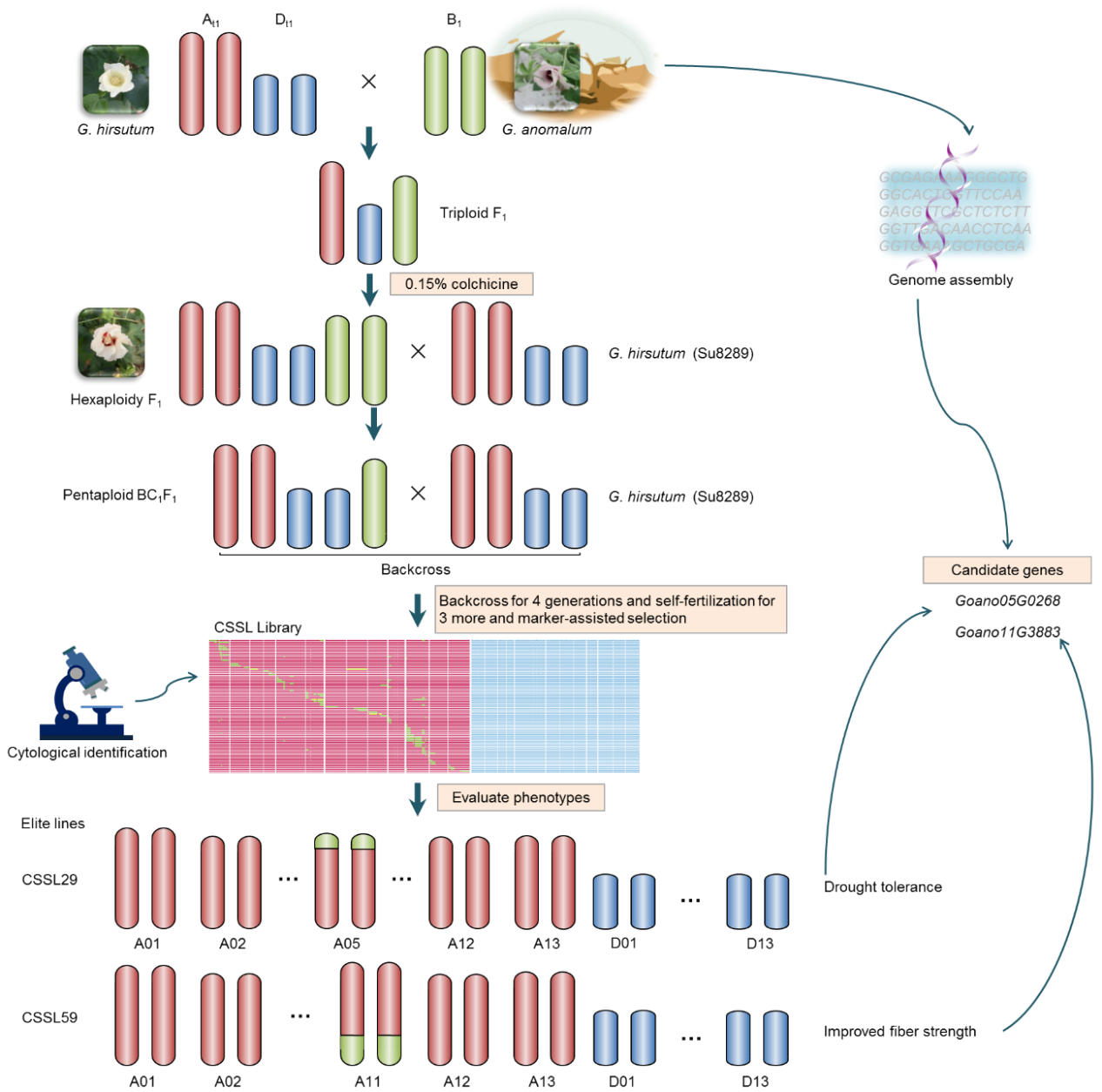
Figure 1. Scheme for generating the CSSL population and rapidly discovering and cloning agronomic genes from G. anomalum.
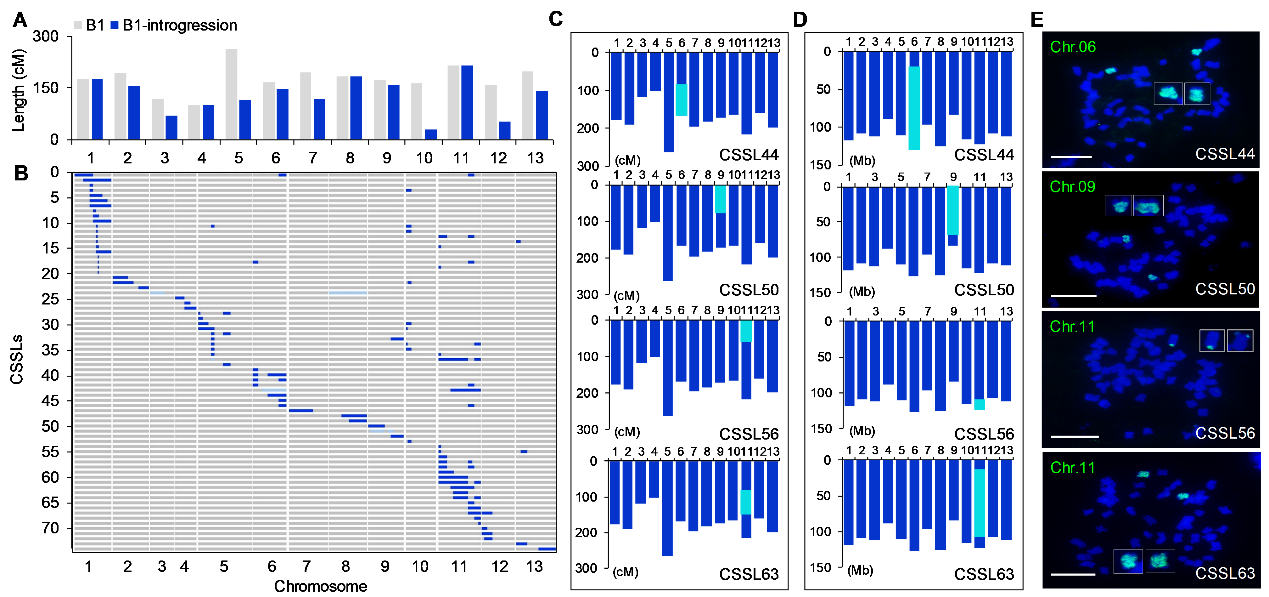
Figure 2. Characterization of the CSSL population derived from G. anomalum based on SSR markers, re-sequencing, and FISH experiments.
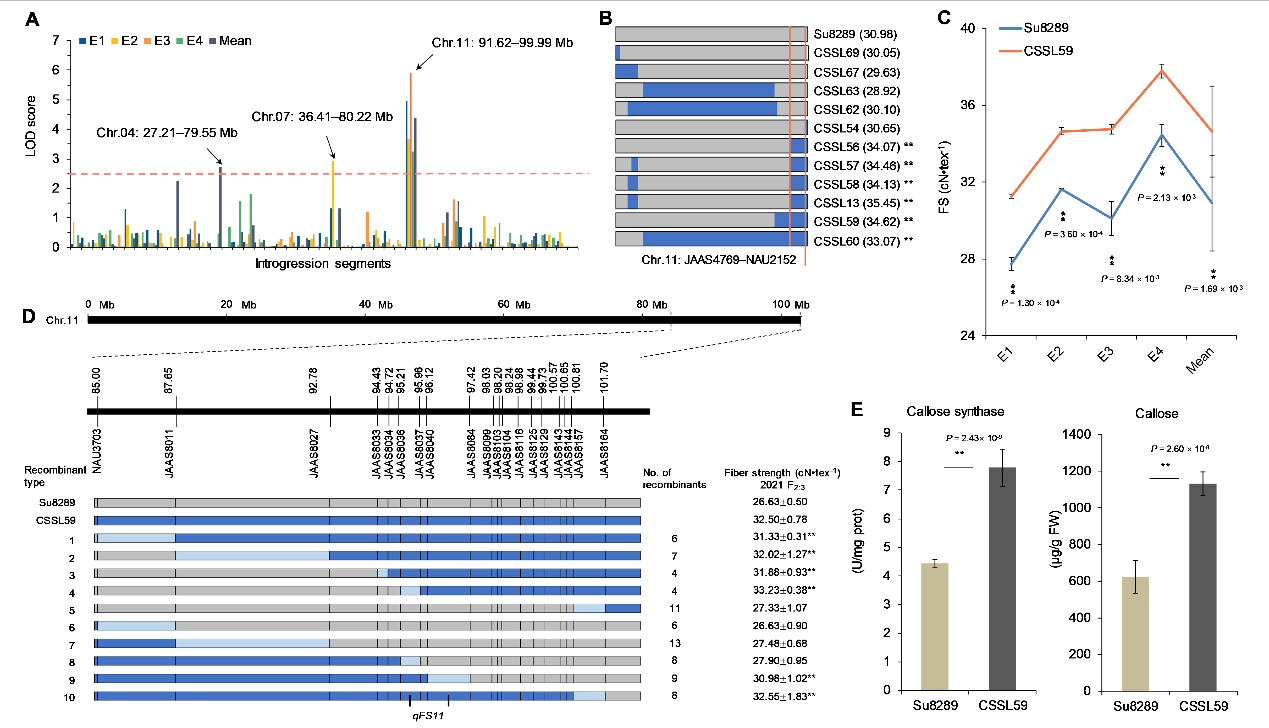
Figure 3. Identification of valuable substitution segment associated with fiber strength and characterization of the candidate gene on Chr.11 of G. anomalum.
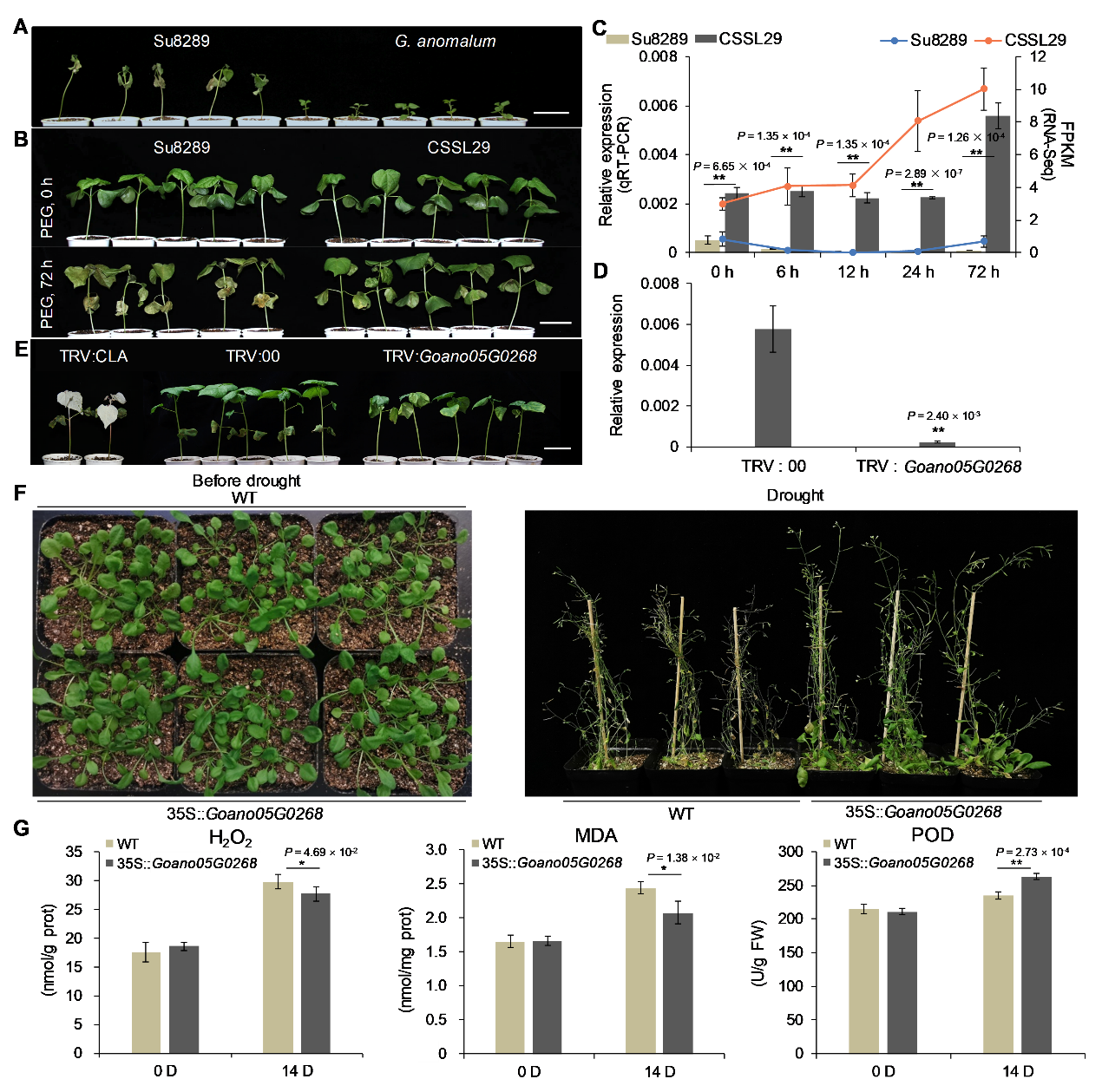
Figure 4. Drought tolerance of CSSL29 and functional verification of Goano05G0268 by VIGS in CSSL29 and ectopic expression in Arabidopsis.
Read more: https://doi.org/10.1016/j.xplc.2022.100350


