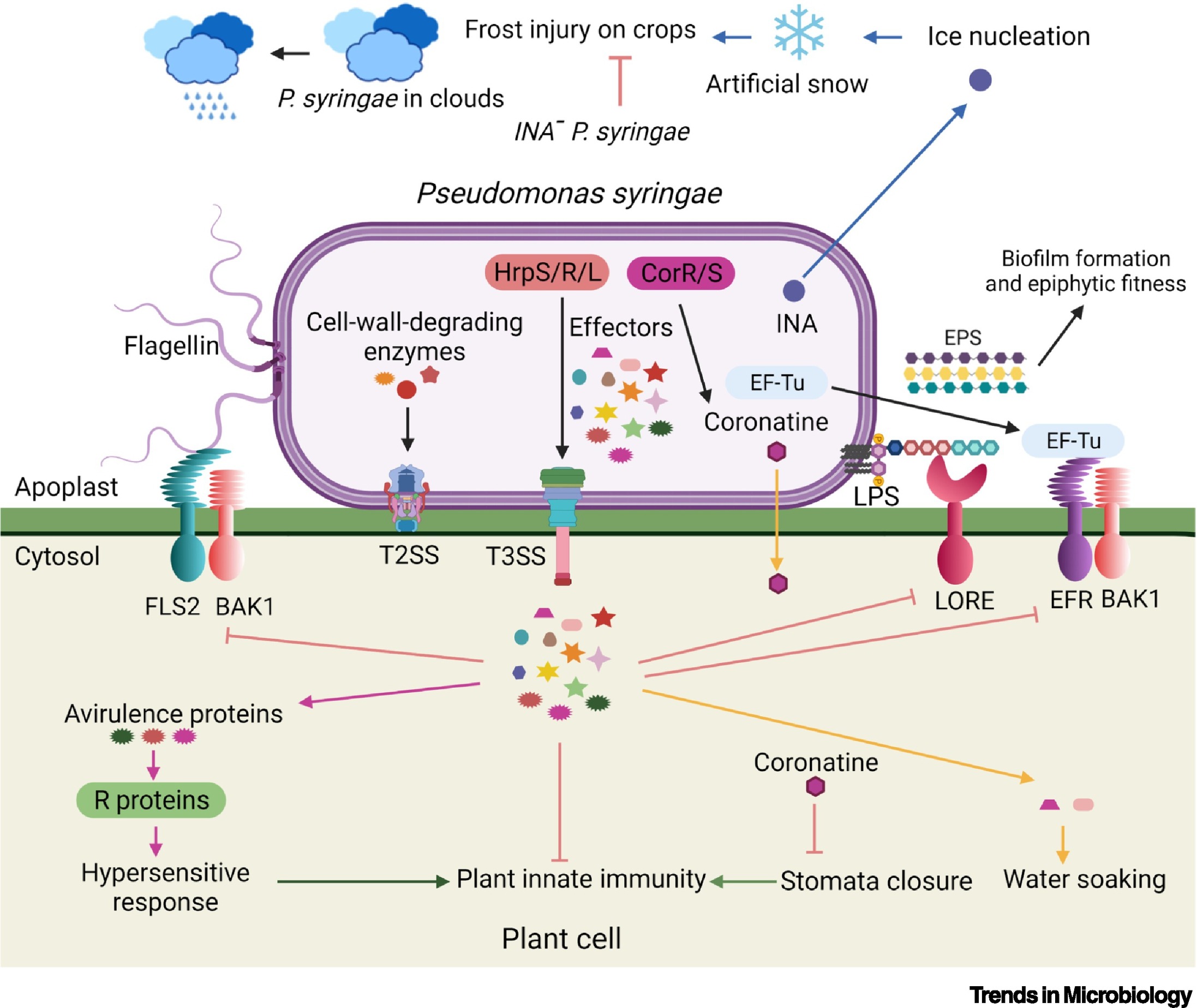
Pseudomonas syringae is one of the best studied plant pathogens. P. syringae pathovars exhibit unique host specificities by infecting different kinds of plants, causing huge economic losses. Studies on the molecular interactions between P. syringae pathovars and plants have made huge contributions towards our understanding of the plant immune system and microbial pathogenicity. Flagellin, lipopolysaccharide (LPS), extracellular polysaccharides (EPS) and EF-Tu from P. syringae are recognized by their receptors/coreceptors FLS2-BAK1, LORE, and EFR-BAK1, respectively, to activate immunity triggered by pathogen-associated molecular patterns (PAMPs). P. syringae relies on cell-wall-degrading enzymes secreted through the type II secretion system (T2SS), type III effectors delivered into plant cells by the type III secretion system (T3SS), and toxins, such as coronatine, to cause diseases. Plants have developed resistance (R) proteins to recognize some of these effectors or avirulence proteins, inducing the hypersensitive response. Ice-nucleation-active (INA) proteins from P. syringae, which can initiate ice formation, have been used to produce artificial snow.

KEY FACTS:
P. syringae was first isolated from the lilac tree (Syringa vulgaris). Due to its production of the siderophore pyoverdin on King’s B medium, P. syringae has a yellow fluorescent appearance.
The complete genome sequence of the first P. syringae, Pseudomonas syringae pv. tomato (Pst) DC3000, was published in 2003. The genome size of Pst DC3000, which has a circular chromosome and two plasmids, is about 6.5 Mb.
Type III effectors (T3Es or Hops) that are delivered into host plant cells by the T3SS are required for P. syringae to cause diseases. The two major functions of T3Es are suppression of plant innate immunity and creation of an aqueous apoplastic space for pathogen growth. HopM1 and AvrE, two major T3Es in the Conserved Effector Locus (CEL) of the pathogenicity island, have been identified as the major causes for the water-soaking symptom during the infection.
The first nucleotide-binding site-leucine-rich repeat (NB-LRR) receptor or R protein RPS2 was identified in the plant arabidopsis (Arabidopsis thaliana). RPS2 detects the cleavage of RIN4 by the T3E or avirulence (avr) protein AvrRpt2 from P. syringae, triggering effector-triggered immunity (ETI) through indirect recognition. The hypersensitive response (HR), a rapid localized programmed cell death, is a hallmark of ETI.
Several phytotoxins, including coronatine, syringolin A, syringomycin, syringopeptin, phaseolotoxin, and tabtoxin, have been found in different P. syringae strains. The best characterized P. syringae toxin is coronatine, which mimics the active form of plant hormone jasmonic acid, jasmonoyl–isoleucine (JA–Ile). Coronatine antagonizes the function of salicylic acid in inducing stomata closure, triggering the reopening of stomata to allow pathogen invasion.
Many strains of P. syringae, but not Pst DC3000, harbor ice-nucleation (INA) genes, which encode INA proteins. Once INA proteins translocate to the surface of the bacteria they can function as nuclei for ice formation. Therefore, the INA proteins have been used to make artificial snow. Conversely, P. syringae INA minus (INA–) mutants have been released into fields to protect crops from frost damage. It has been postulated that P. syringae plays a role in the earth’s water cycle because it is frequently found in precipitates and in the center of hailstones.
DISEASE FACTS:
To date, over 50 pathovars of P. syringae have been discovered. P. syringae can cause diseases in many economically important crops, resulting in billions of dollars loss annually. For example, bacterial canker disease, caused by P. syringae pv. actinidiae, poses serious threats to the kiwifruit industry, leading to huge economic losses.
Pst DC3000 not only infects the economically important crop tomato, it also causes disease in the model plant A. thaliana. Both Pst DC3000 and P. syringae pv. maculicola (Psm) strain ES4326 are frequently used to study the molecular interactions between P. syringae and A. thaliana. Many milestone discoveries in molecular plant–pathogen interactions were made using the P. syringae–A. thaliana pathosystem.
Besides Pst DC3000 and Psm 4326, P. syringae pv. syringae (Pss) B728a, which causes brown spot disease on bean, serves as a model to study epiphytic fitness, while P. syringae pv. phaseolicola (Psp) 1448A, which is a causal agent of halo blight on bean, has been involved in the study of avr genes and non-host resistance.
Read more:https://doi.org/10.1016/j.tim.2022.03.002


