Agrobacterium tumefaciens-mediated transformation (ATMT) is becoming a popular effective system as an insertional mutagenesis tool in filamentous fungi. To gain more insight into the cellular and molecular mechanisms in the pathogenesis of Ustilaginoidea virens, the causal agent of rice false smut disease, a T-DNA insertion mutant library of U. virens was established using ATMT. We optimized a range of conditions to improve the transformation efficiency. Transformants were most effectively obtained when the optimal co-cultivation time is 72 h, with 50 μM AS in medium and 100 μl A. tumefaciens for co-cultivation, leading to the production of 160–185 hygromycin B resistant transformants per 1 × 105 conidia. Southern blot analysis indicated that 58.14% of transformants had a single T-DNA copy. Among 5600 transformants tested for virulence, 37 mutants with reproducible pathogenic defects were obtained. The flanking sequences of three avirulent tranformants (B20, B1015 and B1465) and two pathogenicity-reduced transformants (B726 and B785) were amplified by high-efficiency thermal asymmetric interlaced PCR. Sequence analyses revealed that single T-DNA insertion in mutant B20 targeted the coding region of a gene encoding a protein highly similar to SUN family protein, and in mutant B726 targeted upstream of a gene with unknown function. The two T-DNA insertion sites in mutant B785 were found in the coding region of a gene encoding C6 transcription factor, but failed in amplified flanking sequence of another T-DNA. Chromosomal rearrangement occurred in the genome of mutant B1016 and B1465 with single T-DNA insertion. Among avirulent mutants, B20 showed altered colony growth and pigmentation. The T-DNA insert in B20 was detected in the coding region of a gene named UvSUN2. Morphophysiological characterization analysis suggested that UvSUN2 might be a virulence factor, and possibly required for proper fungal growth, cell wall construction, and stress responses in U. virens. This study optimize and validate the transformation procedure, maximizing the number of single copy transformants and developing an efficient procedure for rescuing adjacent host sequences along with the inserted T-DNA. This is the first report of identification several candidate virulence factors and validated one in U. virens. Together with identification of avirulent mutants and their associated genes, results suggested that ATMT can effectively be used to identify genes associated with pathogenicity in U. virens, and provided novel insights into molecular mechanisms underlying virulence of this pathogen.
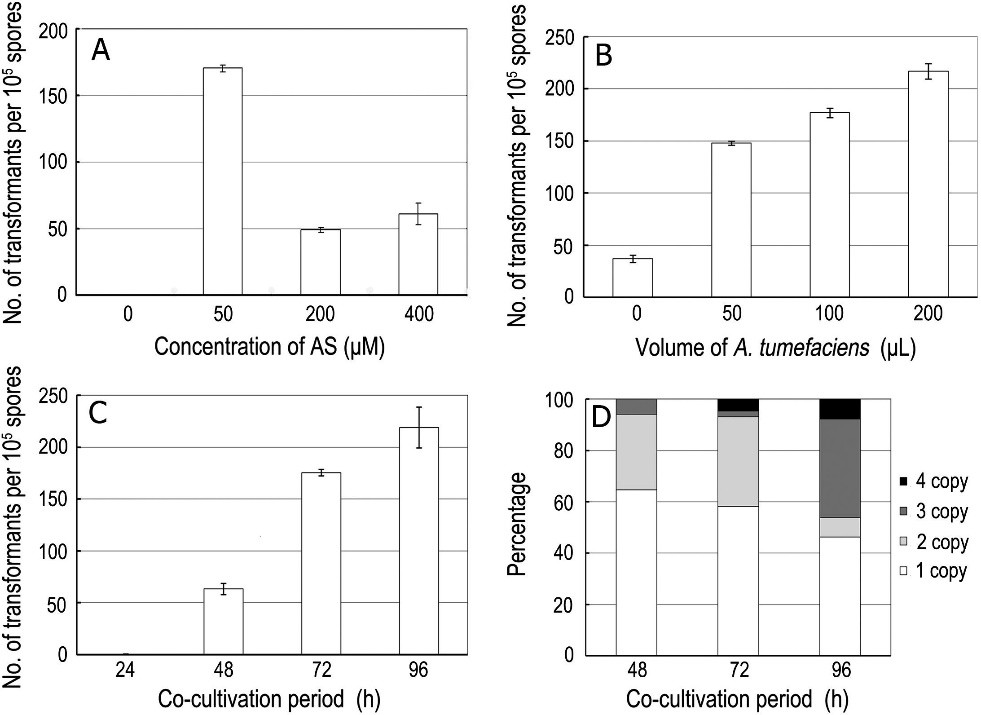
Fig. 1. Effect of concentrations of acetosyringone, volume of A. tumefaciens cells and time of co-cultivation period on transformation efficiency. (A) Effect on transformation efficiency of increasing the concentration of acetosyringone (AS). (B) Effect on transformation efficiency of the volume of A. tumefaciens cells. (C) Effect on transformation efficiency of increasing the co-cultivation time. (D) T-DNA copy number analysis completed with co-cultivation time. Data presented are the average of six plates per treatment. Error bars indicate standard errors.

Fig. 2. Virulence of mutants generated by Agrobacterium tumefaciens-mediated transformation and fluorescence images of Ustilaginoidea virens. (A) The pathogenicity phenotype of transformants on rice of Ustilaginoidea virens 25-day after inoculation. B20, B1016, and B1465 were pathogenicity-deficient transformants; B1178 and B2588 were transformants reduced virulence. (B) Expression of green fluorescent protein in conidia, mycelium, and chlamydospores of Ustilaginoidea virens transformants were analyzed by compound microscope.

Fig. 3. Amino acid sequence alignment and phylogenetic analysis of UvSUN2 with the homologs from other species. (A) Alignment of the predicted amino acid sequence of UvSUN2 with its homologs from Saccharomyces cerevisiae. SUN4, SIM1, UTH1, NCA3, Ymr244w. Identical amino acids are shown on a black background and similar amino acids are shown on a pink background. Red asterisks indicated the cysteines of C-X5-C-X3-C-X24-C motif. Light red showed the multiple insertions in the conserved SUN domain in B20 and Ymr244w. (B) Phylogenetic tree of UvSUN2 and 13 homologs from other species was constructed by observed divergency distance method in the program MEGA 5.1. Numbers at the nodes in the rooted tree represent bootstrapping value on 1000 replications. Numbers correspond to GenBank accession numbers. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
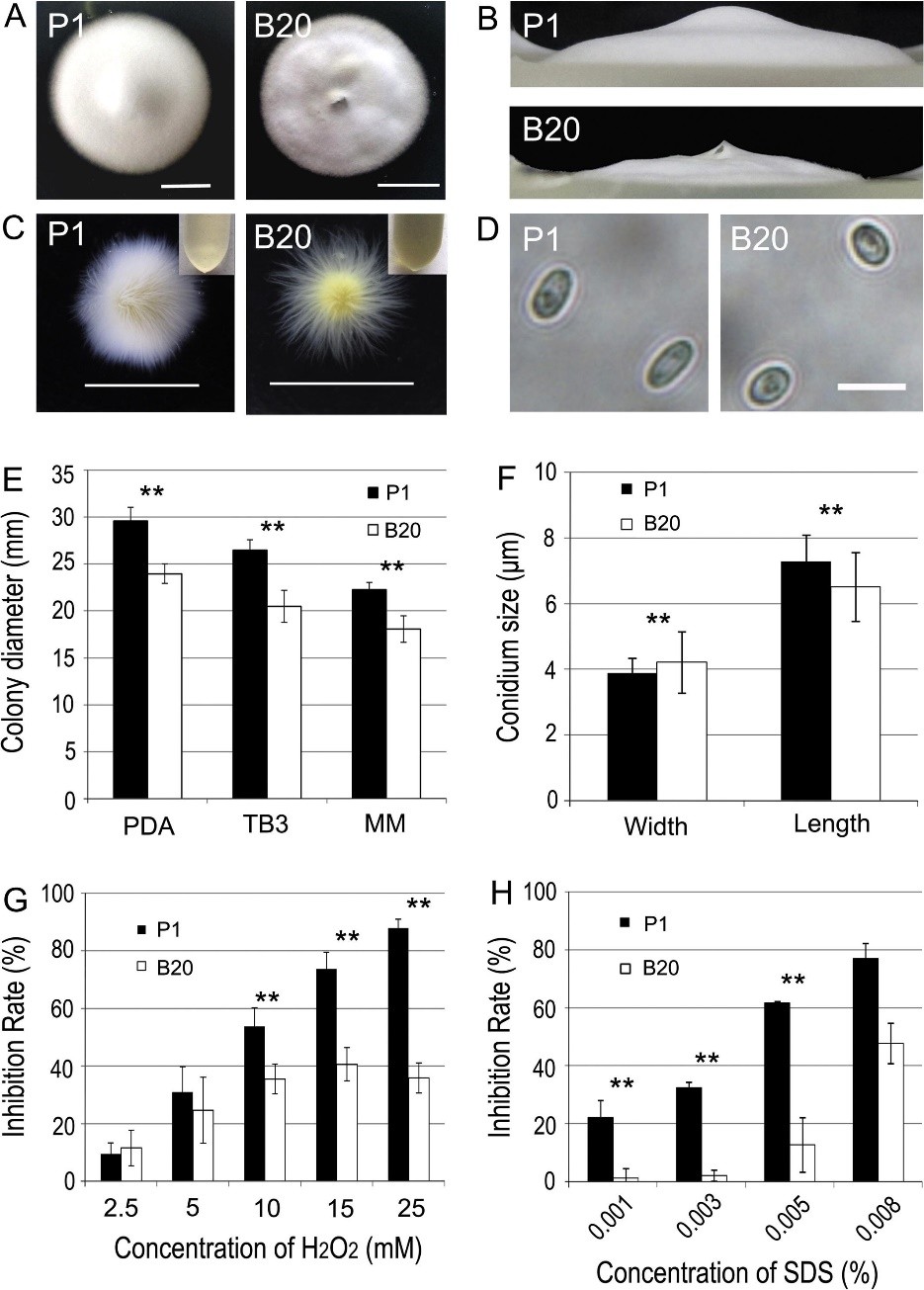
Fig. 4. Phenotypic analysis of B20 mutants of Ustilaginoidea virens. Mycelial growth is altered in the B20 mutant (A and E). The B20 mutants and the wild type strain (P1) were inoculated on PDA medium and cultured at 28 °C in darkness for 12 days. (Bar = 1 cm). (B) Aerial hyphae growth is reduced in B20 mutants on PDA medium for 12 days at 28 °C. (C) Phenotype of mycelia growth in liquid CM medium. The B20 mutants and the wild type strain (P1) were inoculated on PDB medium and cultured at 28 °C in darkness for 5 days with shaking at 150 rpm. The culture medium supernatant liquid was shown at the top right corner. Bars equal 1 cm. (D) Conidium morphology. Conidia were harvested from PDB medium, and observed by light microscopy. (Bar = 10 mm). (F) Conidia size comparison. (G) The colony diameters of the testing strains were measured under oxidative stress and subjected to statistical analysis. (H) The colony diameters of the testing strains were measured under SDS stress and subjected to statistical analysis. The growth inhibition rate is relative to the growth rate of each untreated control [inhibition rate = (the diameter of untreated strain – the diameter of treated strain)/(the diameter of untreated strain ×100%)]. Three repeats were performed and similar results obtained. Asterisks indicate that the difference is statistically significant. Error bars represent standard deviations. Double asterisk indicates that the differences between the transformant B20 and the wide type strain P1 are significant (p < 0.01).


