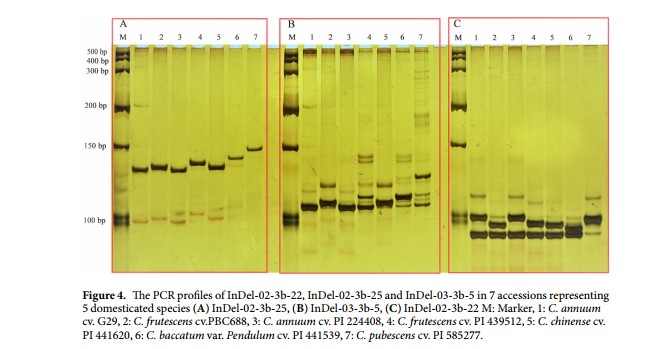
Genome-wide identification of Insertion/Deletion polymorphisms (InDels) in Capsicum spp. was performed through comparing whole-genome re-sequencing data from two Capsicum accessions, C. annuum cv. G29 and C. frutescens cv. PBC688, with the reference genome sequence of C. annuum cv. CM334. In total, we identified 1,664,770 InDels between CM334 and PBC688, 533,523 between CM334 and G29, and 1,651,856 between PBC688 and G29. From these InDels, 1605 markers of 3-49 bp in length difference between PBC688 and G29 were selected for experimental validation: 1262 (78.6%) showed polymorphisms, 90 (5.6%) failed to amplify, and 298 (18.6%) were monomorphic. For further validation of these InDels, 288 markers were screened across five accessions representing five domesticated species. Of these assayed markers, 194 (67.4%) were polymorphic, 87 (30.2%) monomorphic and 7 (2.4%) failed. We developed three interspecific InDels, which associated with three genes and showed specific amplification in five domesticated species and clearly differentiated the interspecific hybrids. Thus, our novel PCR-based InDel markers provide high application value in germplasm classification, genetic research and marker-assisted breeding in Capsicum species.