As one of the important vegetable, industrial, and model crops, the tomato (Solanum lycopersicum) genome had been sequenced in 2012 (The Tomato Genome Consortium, Nature). One major finding is that the tomato genome is composed of many kinds of repetitive sequences, particularly transposable elements (TEs). To further investigate the structural characteristics, distribution pattern, and evolutionary process, we annotated and reanalyzed the TEs in tomato plants. The current data showed that the relatively old long terminal repeat retrotransposons (LTR-retrotransposons) are mainly located in gene-poor heterochromatins, and in contrast, the relatively young LTR-retrotransposons are enriched in gene-rich euchromatins. Further analysis indicated that such distribution patterns might be caused by the suppression of activities of TEs in heterochromatins. Since the distribution of TEs in tomato plants are different from those reported in other plant genomes, the genome evolution of tomato may be specific after split with its relatives from their common ancestor.
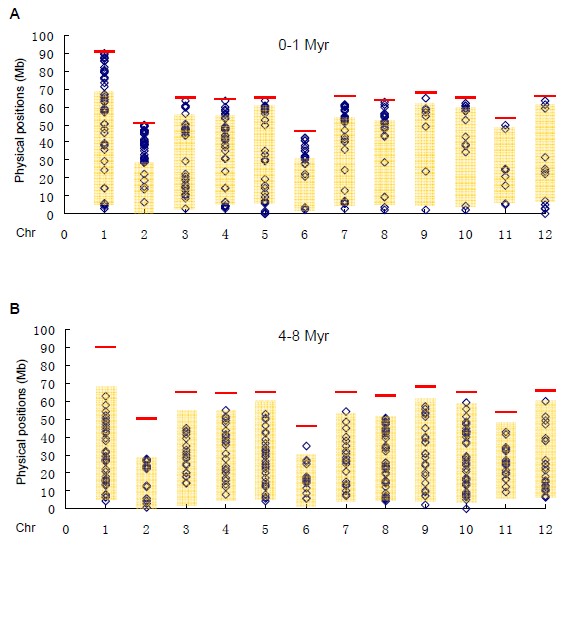
Figure 1.1 The distributions of relatively young (A) and old (B) LTR-retrotransposons along 12 tomato chromosomes.
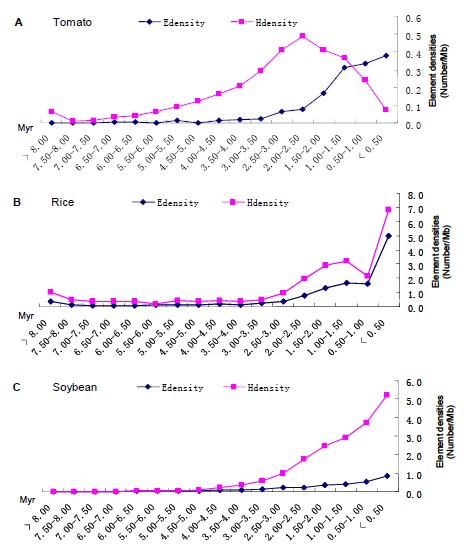
Figure 1.2 Comparisons of the distributions of LTR-retrotransposons in tomato (A), rice (B) and soybean (C).


